Building a secure and performative DICOM viewer
Client Background
Our client is a Professor of Neuro-Oncology leading a research group focused on brain tumor monitoring. The team works with MRI/CT studies in DICOM format and needs reliable tools for longitudinal review and research-grade annotations. Their goal: speed up manual work, improve data quality, and create privacy-safe datasets for machine learning.
Business challenge
Existing viewers were not a fit. They lacked strong tools for timepoint comparison, had weak de-identification, or made data export for AI training painful. The client asked us to build a DICOM viewer that can:
- Load studies from local files and remote data storages.
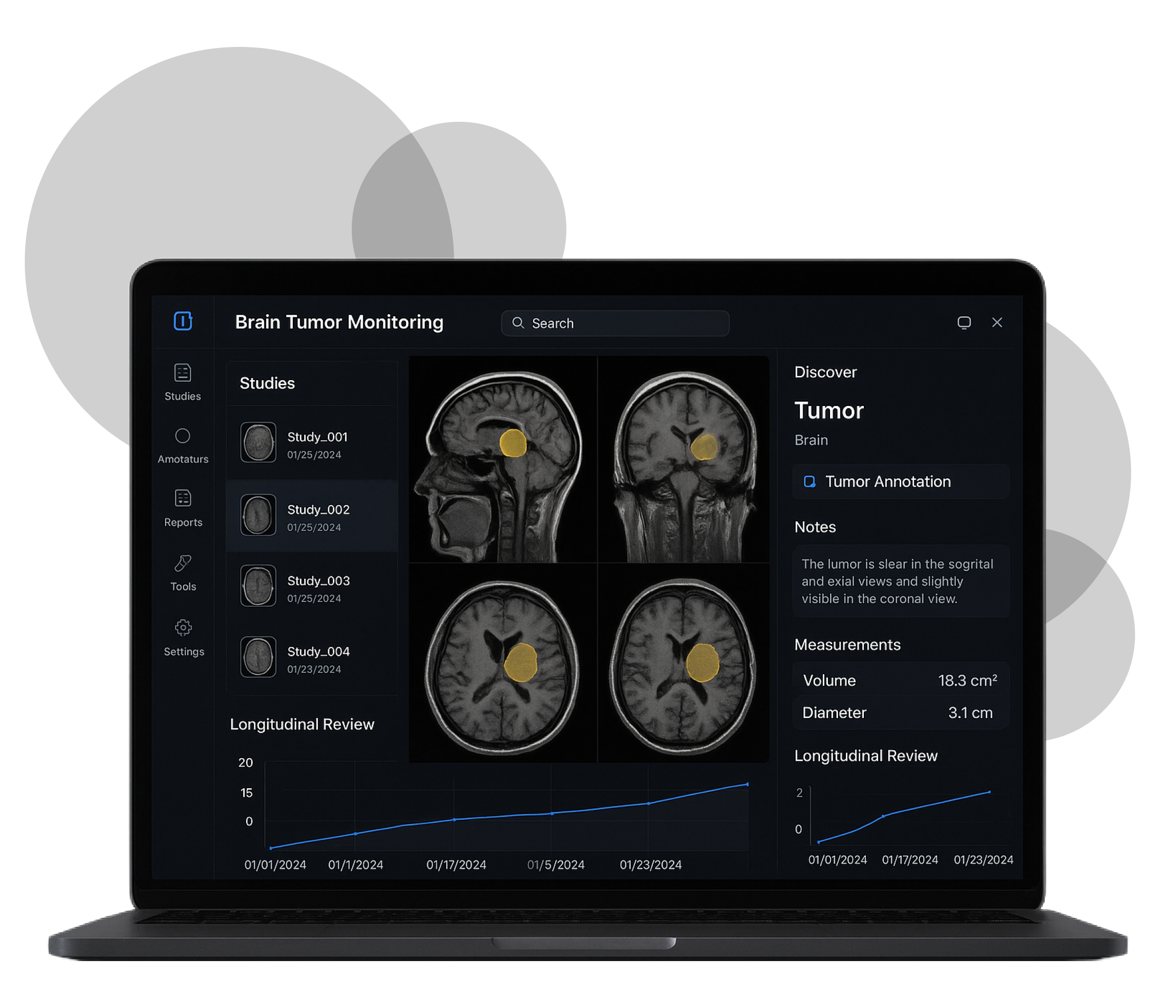
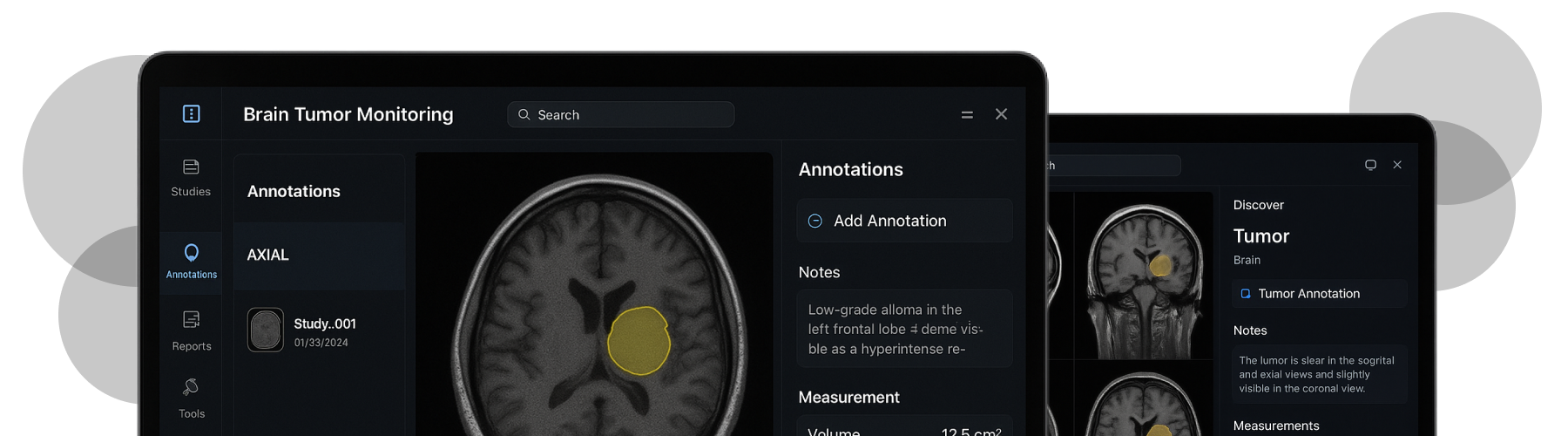
- Enable precise manual contouring across slices and timepoints.
- Compute 3D tumor volumes from 2D contours.
- Remove PHI before export.
- Export clean data for downstream ML pipelines.
How did we make it work?
We designed a solution with a pluggable data layer and a privacy-by-design pipeline.
- Data ingestion: Local upload, DICOM over TCP/IP and DICOMweb images retrieval.
- Viewer UX: Fast slice navigation, window/level presets, series comparison, consistent hotkeys.
- Annotation & Volume: 2D contours per slice with voxel aggregation into 3D volume; versioning and audit trail.
- De-identification (PVI scanner): Removes private tags and burn-in text before export; configurable rules.
- AI-ready export: DICOM SEG and NIfTI + JSON with stable pseudo-IDs for longitudinal linking.
- Governance: Role-based access, activity logs, encryption in transit/at rest.
Main technical challenges of the DICOM web viewer
01 Longitudinal alignment across timepoints

Tumor comparison requires consistent orientation and series matching. We built utilities for baseline vs. follow-up alignment and kept stable pseudo-identifiers after anonymization.
02 High-precision manual annotation in the browser

Accurate contouring is hard on large studies. We optimized the canvas/OpenGL path, added snap/adjust controls, and ensured low edit latency even on high-slice counts.
03 Reliable 3D volume computation

Errors compound across slices. We validated voxel spacing, slice thickness, and interpolation to produce trustworthy volume totals, with change tracking between sessions.
04 Robust de-identification

PHI hides in both headers and images. Our PVI scanner strips private tags and detects/removes burn-in overlays before any export leaves the system.
05 DICOMweb integration and performance

Remote fetches can be slow and inconsistent. We implemented range requests, smart caching, and progressive loading to keep the UI responsive during large downloads.
Value delivered by devPulse
- Purpose-built workflow for neuro-oncology
Slice-by-slice contouring, 3D volume, and timepoint comparison in a single, browser-based tool.
- Privacy-safe data sharing
Zero-PHI exports (headers + burn-ins removed) simplify IRB and cross-team collaboration.
- AI-ready datasets
One-click export to DICOM SEG / NIfTI + JSON enables fast downstream training and evaluation.
- Traceability and quality
Versioned annotations, audit trails, and configurable reviewer workflows.
Development Timeline
2019-2020
Team Composition
- 3 Developers
- 1 QA Engineers
- 1 Product Manager
Target Platforms & Tech
- Windows
- Mac
- DICOMweb, DICOM over TCP/IP
- OpenGL, VTK
- DCMTK, GDCM
- QT, QML
- CMake
Location
USA
Methodology
Use from above

Anna Tukhtarova
CTO devPulse
A technology-driven leader with over 15 years of deep expertise in architecting and delivering complex enterprise solutions that operate seamlessly across cloud, on-premises, native, and embedded environments.
Request A Free No-Obligation Quote
By clicking "Send A Message", You agree to devPulse's Terms of Use and Cookie Policy.





